Q:
we are working on lncRNA. We have learned a lot from you publication
“Whole-genome analysis of papillary kidney cancer finds significant
noncoding alterations” published on PLoS Genet. And we want to get more
detailed information about this study from you. Would like to tell us the
detailed final nucleotide mutated and mutation frequency in NEAT1 and
MALAT1?
If you were kindly offering us this information, it would be very helpful
for us.
A:
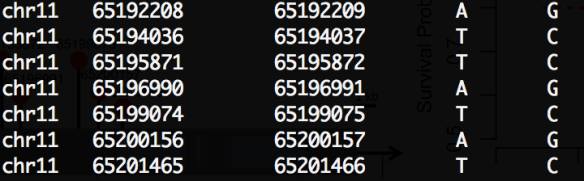
Below is the detailed mutation information in NEAT1 (BED format, hg19). The second and fifth mutations are in the same sample. The cohort size is 35.