Q1:
First of all I must show great respect to your brilliant work on developing the PseudoPipe software.
Now I am working on my graduate paper, and need to use this software. But I met some problems, so any guide or assistance from you would be appreciated.
I just download the software package from your website and unpack it in my home directory(that is ~/), but when I test it according to your manual, it reported errors as below:
I have tried several ways to fix it ,even trying to modify the source code, but failed. I’ve been driven somehow crazy haha.
Can you please provide some suggestions? thanks in advance!
A1:
It looks like your installation is not referencing python properly. Please edit the env.sh file with the appropriate source/path for python in your system.
Q2:
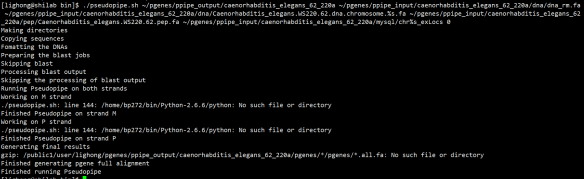
According to your suggestion, now I have finished all the environment variable setting in env.sh, but I still got error while running the software(as the below Fig.1)
So I try to fix the code of pseudopipe.sh , and I finally made it run just by modifying the "source setenvPipelineVars" into "source ./setenvPipelineVars" at line 141. And I got the final result file(as Fig. 2) by running your sample data. Is the result correct?
Don’t know if anybody reported similar error before. If not, I hope it would contribute to improving your powerful software. And it would be great if you can also display on your manual or README what the standard output and final result file look like when testing the sample data.
A2:
The results look right. Thank you for your suggestions, we will take them into account in a future update of the pipeline.
Good luck with your analysis.